Posters
Publications
Product Manuals
Safety Data Sheets
FAQs
General FAQs
Does the ProMTag permanently modify tagged proteins?
No! The ProMTag linkage is completely reversible and proteins are left in their original, unmodified state following ProMTag reversal.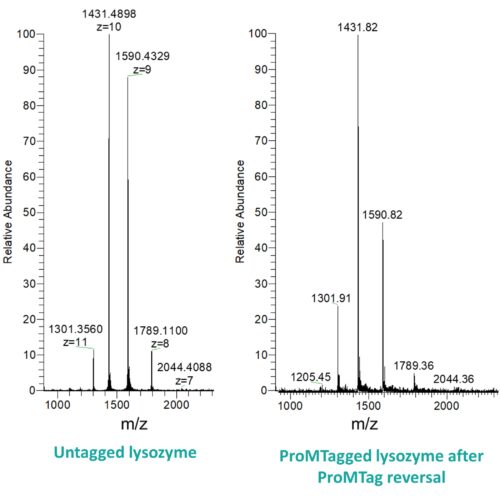
What are RC tubes and how do I use them?
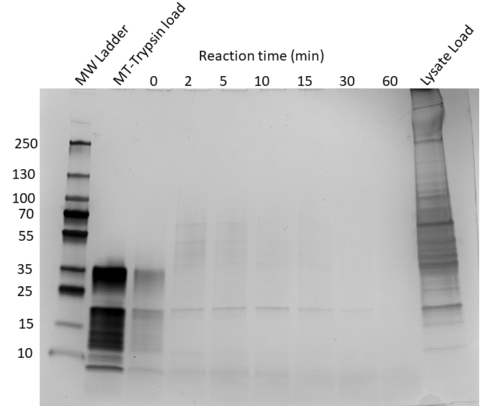
Resin Capture (RC) tubes are designed with a small slit in the bottom that allows for the passing of liquid with very little dead volume, but retains the solid resin. The RC tubes have two main features: a slit at the bottom of the tube, and a vent hole at the top. These tubes function similarly to a typical spin column, but require only brief (<5 seconds) centrifugation steps to fully spin out the liquid. When using the RC tube, take care to avoid touching the slit, and always spin and vortex the RC tube in an adapter tube.What is special about MT-Trypsin?
MT-Trypsin is a methyltetrazine (MT) labeled trypsin that is stabilized, resistant to autolysis, and capturable by TCO resin. This allows for short digestion times (1 hour) at high MT-Trypsin concentration without the risk of overwhelming the final sample with trypsin autolysis peptides. During a 1 hour digestion period, MT-Trypsin will completely digest input proteins and also be captured on the TCO resin.